
Video
Autophagy - Macroautophagy \u0026 Importance in HealthAutophagy and mTOR signaling -
Many components of the autophagic machinery and autophagy receptors such as p62 present in these organelles are themselves substrates of the process being subjected to lysosomal degradation However, Gαq is not a substrate of autophagy under nutrient stress conditions, since serum removal did not promote a significant decrease of Gαq protein levels in WT MEFs, or in cells deficient for Atg5, a well-established model of macroautophagy deficiency, contrary to what was observed for the known autophagy substrate p62 Supplementary Fig.
Overall, the unexpected location of Gαq in autophagic compartments and lysosomes and the fact that this protein is not degraded under nutrient stress conditions in these organelles suggested that the observed changes in autophagy upon Gαq modulation may result from a regulatory function of this protein on autophagy.
A representative blot of four independent experiments is shown. A representative blot of five independent experiments is shown.
For a and b , autophagic markers LC3-II and p62 and the activation levels of the mTORC1 and AMPK pathways were analyzed by assessing the phosphorylation status of the downstream target of mTORC1 S6 ribosomal protein see Supplementary Fig.
Tubulin and GAPDH for the p-AMPK and AMPK blots in panel a were used as loading controls. The main cellular nutrient sensor pathways converge in the AMPK and mTORC1 cascades to regulate metabolism and autophagy.
The expected attenuation of the mTORC1 cascade in WT cells upon low serum Fig. Of note, in serum-starved cells where downmodulation of mTORC1 pathway and upregulation of autophagy takes place Fig. a Experiment outline diagram. Tubulin, actin, or GAPDH were used as loading controls. Blots are representative of three panels b , d , and panel e lower blots or five panels c and e independent experiments.
See Supplementary Fig. In control cells without CNO stimulation , this pattern persists in the subsequent hours, as assessed by the decreased status of p-S6 Fig. A similar pattern of autophagy modulation was detected when testing the evolution of p62 levels Supplementary Fig.
Once again, we did not detect changes in the AMPK pathway. It is worth noting that the reactivation of the mTORC1 pathway triggered by CNO does not completely parallel the pattern of Akt activation Fig. These data were consistent with the occurrence of alternative routes of Gq-mediated mTORC1 stimulation.
CNO-mediated preservation of mTORC1 activation also attenuated the increase in LC3-II levels triggered by amino acid removal in control cells Fig. Consistent with this notion, overexpression of constitutively active mutants of Gαq Gq-RC and Gq-QL was able to maintain mTORC1 pathway activation even under low-serum conditions Supplementary Fig.
This mTORC1 reactivation is essential to inactivate ongoing autophagy and return to basal conditions. Two different refeeding paradigms were used. WT vs.
Tubulin was used as a loading control. Representative blots are shown. a , b Statistical significance was analyzed using two-sided unpaired t -test. Image on the right shows the quantification analysis of lysosome distribution.
Representative blots of three independent experiments are shown. Control vs. Immunofluorescence with LAMP1 to highlight the endolysosomal compartment revealed that WT MEFs displayed the expected switch of lysosomes from the cell periphery to a more perinuclear location upon exposure to amino acid-free medium 25 , whereas the re-exposure to amino acids restored their original location Fig.
These results are consistent with their inability to stimulate mTORC1 in response to nutrient recovery. Intriguingly, immunoprecipitation of either endogenous Gαq Fig. Importantly, immunoprecipitation of endogenous Raptor, a specific component of mTORC1, also revealed the presence in the same complex of Gαq, p62, and mTOR Fig.
The mTORC1 activation complex formation undergoes a dynamic modulation upon nutrient fluctuations 15 , Following amino acid deprivation, we found dissociation of p62 from mTOR, while Raptor and Gαq remained in the mTOR complex Fig.
In response to amino acid replenishment, p62 re-associated to the mTORC1 complex. Blots are representative of three a , c and four b independent experiments.
c WT vs. d Control vs. We searched for potential molecular links between Gαq and mTORC1 activation. A novel acidic region in Gαq responsible for its direct interaction with the PB1-containing protein Protein kinase C ζ PKCζ has been reported, and glutamic acid residues and proven to be critical for such interaction Remarkably, the stimulation of the mTORC1 pathway triggered by transient expression of Gαq WT in low-serum conditions was not mimicked upon overexpression of the Gq-EEAA mutant Fig.
Overall, these results suggested that the PB1-binding region of Gαq is involved in mTORC1 pathway modulation. A representative blot of three independent experiments is shown.
c — f CHO cells were transiently transfected with the indicated combinations of HA-p62, Gαq WT or the indicated Gαq mutants constitutively active Gq-QL or GqRC, active but PLC-β activation-defective GqQL-AA, defective in binding to PB1-domain-containing proteins Gq-EEAA and constitutively active GqRC-EEAA , or the PB1 domain of p62 GFP-Flag-PB1-p Gαq WT vs.
We, therefore, used transient overexpression of Gαq and HA-p62 constructs to characterize their association. Of note, p62 displays features of a bona fide Gαq effector reviewed in ref. Consistent with data in Fig.
Of note, Gαq mutants Gαq-YF, Gαq-TE, Gαq-WD unable to bind GRK2 co-immunoprecipitated with p62 to an even greater extent than wild-type Gαq Supplementary Fig. Consistent with a PB1-like association between Gαq and p62, the overexpression of the PB1 domain of p62 clearly inhibited the complex formation Fig.
a CHO cells were transfected with HA-p62 and Gαq constructs, starved with 0. c Basal activation status of the mTORC1 cascade and autophagy levels correlates with the ability of Gαq mutants to interact with p mTOR immunoprecipitates and total lysates were analyzed by western blot with specific antibodies to determine the components of the complex and overall cell expression levels, respectively.
The blots shown are representative of three independent experiments. e mTORC1 reactivation and autophagy modulation in response to amino acid recovery after starvation correlate with the ability of Gαq to interact with p Statistical significance was analyzed using two-way ANOVA with Bonferroni multiple comparison test.
Basal activation status of the mTORC1 cascade and autophagy levels correlated with the ability of Gαq mutants to interact with p Overall, these results support the notion that the interaction between Gαq and p62 in response to nutrients is key for the stimulation of the mTORC1 pathway and the control of autophagy.
Our data suggest that Gαq acts as a general and relevant modulator of mTORC1 signaling over autophagy in response to fluctuations in different types of nutrients. The metabolic modulation of autophagy is mainly orchestrated by the balance between the opposite inputs provided by the mTORC1 negative and AMPK positive pathways 12 , 31 , Moreover, constitutively active mutations in Gαq maintain mTORC1 activated in absence of serum.
Fifth, consistent with its role as an autophagy modulator, we demonstrate the presence of an endogenous Gαq pool in autophagic compartments and lysosomes in rat liver, where it co-localizes with mTOR, Raptor, and p62, and immunoprecipitation of endogenous mTOR or Raptor from WT MEFs in the presence of nutrients shows that Gαq is part of the mTORC1 multimolecular complex.
This signaling adaptor contains several functional regions and displays a variety of subcellular locations, including autophagosomes and lysosomes reviewed in refs.
p62 plays a central role as an autophagy receptor by forming PB1-linked homo-oligomers, recruiting ubiquitinated cargos, and inducing nucleation of the autophagosome membrane via its interaction with LC3 33 being itself a substrate of autophagy p62 is also involved in other cellular processes 33 , including the modulation of mTORC1 see below.
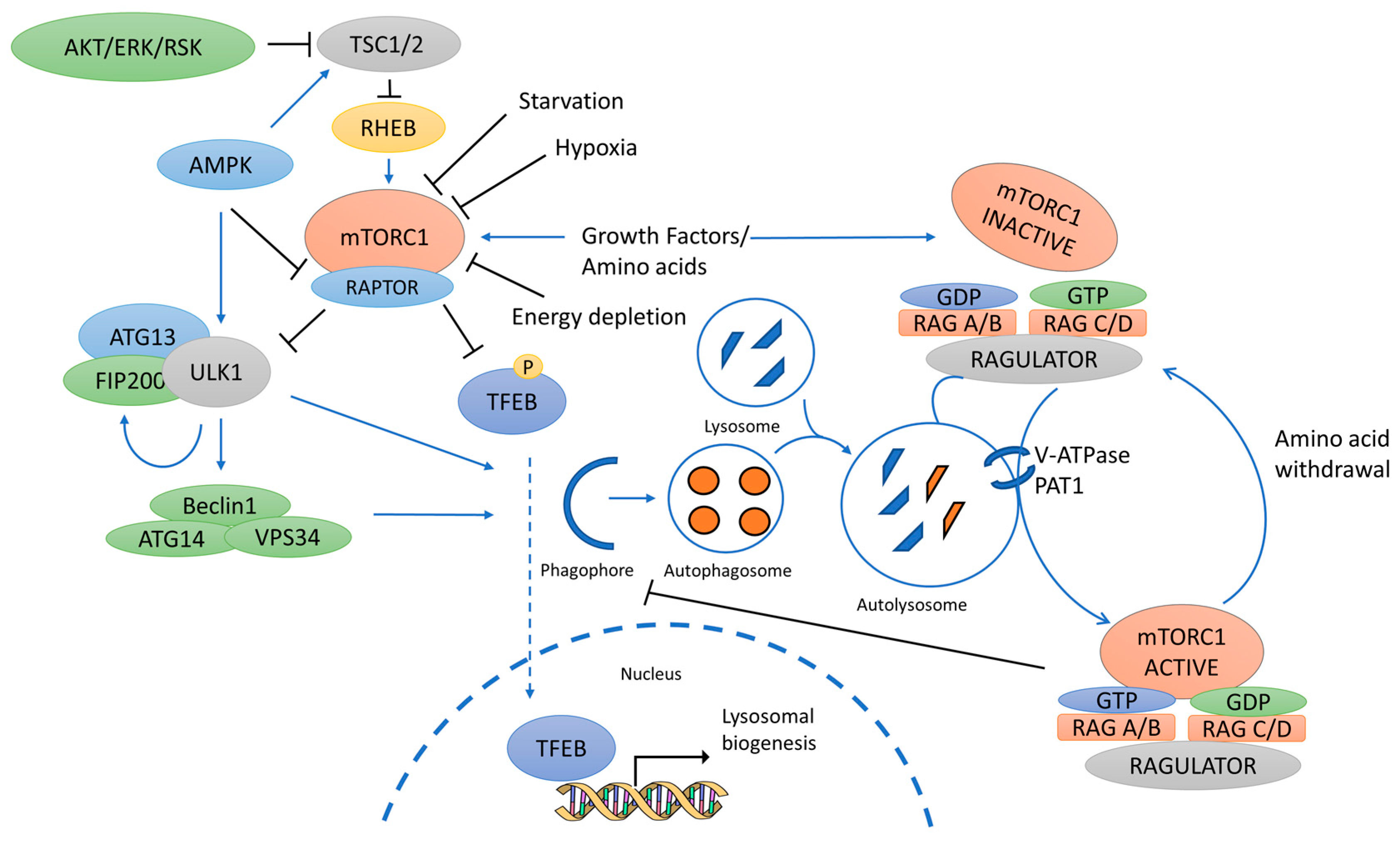
The mTORC1 signaling hub displays an intricate spatiotemporal regulation, involving both upstream signaling pathways and timely recruitment to lysosomes 12 , Major components of the mTORC1 complex include mTOR kinase, mLST8, DEPTOR, and the specific regulatory protein Raptor, which plays a relevant role in facilitating substrate recruitment and in controlling mTORC1 subcellular localization 12 , Importantly, the presence of amino acids, acting through different sensors, is key for full mTORC1 activation by facilitating its recruitment to the lysosomal membrane.
This is achieved via the Ras-related GTP-binding protein Rag family of small GTPases. Glucose-dependent mTORC1 stimulation is also partially regulated through Rag-GTPases by complex mechanisms reviewed in refs. Thus, Rags appear to integrate multiple nutrient inputs converging in mTORC1 modulation.
Importantly, in the presence of amino acids, p62 has been reported to interact with Raptor, thus helping mTORC1 complex translocation to the lysosome The presence of p62 in the mTORC1 multimolecular complex might also contribute to the full activation of kinase activity via TRAF6-mediated K63 ubiquitination of mTOR First, p62, mTOR, Raptor, and Gαq are present in the same multimolecular complex in endogenous conditions in the presence of nutrients, whereas cells lacking Gαq display a reduced presence of p62 in mTOR and Raptor immunoprecipitates along with decreased mTORC1 pathway activation.
Gαq and p62 appear to associate through a PB1-like interaction, involving the PB1 domain of p62 and the acidic PB1-binding region of Gαq, different from the specific interface involved in the association of Gαq with the canonical PLCβ or p63RhoGEF effectors This acidic Gαq region has been shown to interact with the PB1 domain of PKCζ 29 or with the cold-activated channel TRPM8 42 , consistent with this Gαq region being a functional module.
Such core Gαq scaffolding role would be consistent with its general role in autophagy control in response to different types of nutrient stress see scheme in Fig. In nutrient-sufficiency conditions, the presence of serum, amino acids, or glucose would foster the PB1-like interaction between Gαq and p62, leading to basal mTORC1 stimulation and preservation of low, homeostatic autophagy levels.
Upper part: In nutrient-sufficiency conditions, the presence of serum, amino acids, or glucose would foster the PB1-like interaction between Gαq and p62, leading to basal mTORC1 stimulation and preservation of homeostatic autophagy levels.
Lower part: Autophagy levels diagram. In response to starvation, there is a controlled and transient upregulation of autophagy, with a subsequent return to basal levels upon the partial recovery of nutrients. An increasing number of nutrients and metabolites are being identified as endogenous ligands for Gq-coupled GPCR 16 , 17 , On the other hand, relevant serum components as lysophosphatidic acid or sphingosinephosphate and factors as insulin or IGF-1 have been reported to directly stimulate Gαq signaling The presence of Gαq in lysosomal and autophagy-related organelles may be subsequent to receptor-mediated activation at the plasma membrane or represent a stable pool able to undergo nutrient-mediated stimulation at such locations.
Recent data indicate the presence of Gαq in endosomes and other organelles, such as mitochondria 49 , The agonist-dependent presence of GPCR and heterotrimeric G proteins in endosomes as a consequence of GPCR recycling or degradation mechanisms has also been reported Since endocytosis-derived organelles, can form elongation membranes to finally form autophagosomes in response to starvation 54 , endosomes might constitute a source for Gαq in autophagy-related organelles.
Alternatively, activation of Gαq by nutrient-sensitive GPCR could take place directly at locations different from the plasma membrane via molecules that can gain access to the intracellular milieu via specific transporters, as is the case for amino acids, glucose, and other nutrients.
In sum, our results postulate Gαq as a key hub in the autophagy machinery. All rats were housed under pathogen-free conditions and handled according to protocols approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine New York, USA.
Since for these experiments mice were not treated with tamoxifen, we refer to them as WT mice as they display endogenous Gαq levels. We did not use randomization in our animal studies.
We were not blinded to the group in our animal studies. Offermanns Max-Planck-Institute for Heart and Lung Research, Germany. Atg5 WT and Atg5 KO MEFs were provided by Dr.
Mizushima University of Tokyo, Tokyo, Japan and DREADD-Gq-HEK cells human female by Dr. Silvio Gutkind University of San Diego, California, USA CHO cells overexpressing the muscarinic M3 acetylcholine receptor CHO-M3 mice female were a kind gift from Dr.
Tobin University of Glasgow, UK. Cell lines were authenticated by short tandem repeat STR or DNA barcoding analysis and tested for mycoplasma contamination. Human umbilical vein endothelial cells HUVEC human male were grown in Medium Life Technologies, without NaHCO 3 supplemented with 2.
Explants were homogenized using metal beads in a Tissue Lyser Qiagen, Hilden, Germany 60 , and lysates used for immunoblot analysis of mTORC1 pathway modulation. For autophagy modulation by nutrient depletion assays, cells were maintained in 0. Nutrient recovery experiments in MEF cells were carried out by adding increasing percentages of bovine serum or a full amino acid mix MEM amino acids solution 50× Sigma-Aldrich.
For degradation assays, cells were treated with the lysosomal inhibitors mentioned and the proteasomal inhibitor MG 0. The Gαq and Gαq-RC cDNA were from our laboratories Dr. Aragay, CSIC, Barcelona, Spain. Lin Stony Brook University, New York, USA. The Gαq mutant proteins that lack the ability to interact with GRK2 Gαq-TE, WD, YF were a gift from Dr.
Kozasa The University of Tokyo, Japan. GRK2, GRK2-KR, GRK2-DA cDNAs were a gift from Dr. Benovic The Kimmel Cancer Center, USA. The GRK2-RH domain cDNA 61 was generated in our laboratory. The HA-p62 construct was provided by Dr.
Campbell University of Illinois at Chicago, USA. GFP-Flag-PB1-p62 was provided by Dr. Berk University of Rochester, NY, USA. mCherry-GFP-LC3 Addgene, Dr.
Mizushima, constructed by T. Kaizuka University of Tokyo, Japan. cDNAs for lentiviral construction pMD2.
G, pSD. Iggo University of St Andrews, Edinburgh, UK. Gq-wt and psd Gq-EEAA were created in our laboratory from psd GqQL Addgene, Dr. All primers' information has been included in Supplementary Table 1. Gq-EEAA DNA constructs were transfected together with psPAX2 and pMD2. All generated MEF cells lines, psd.
Cells were seeded in a six-well plate a day before transduction. Then, a 2× polybrene mixture was prepared by adding 0. Depending on the cell type these stable cell lines were used until passage 3—4.
Immunoprecipitation was performed with agarose-conjugated anti-HA antibodies Santa Cruz, F-7, scAC. Endogenous immunoprecipitation assays were performed by incubating 0.
Protein concentration was measured using the DC Protein Assay Reagents A, B, and S Bio-Rad. Immunoreactivity bands were quantified by laser densitometry with a Biorad GS scanner and using de Bio-Rad provided Image Lab 5. Vertical lines in western blots indicate juxtaposed lanes that come from the same gel but were nonadjacent.
Cells were mounted with Fluoromount-G-Dapi Southern Biotech. Sample images were acquired using a confocal laser microscope LSM Zeiss at ×63 magnification. Image J software NIH was used for general image analysis and manipulations. Imaging of the mCherry-GPF-LC3 reporter was carried out using high-content microscopy.
The total number of autophagic vacuoles AV, autophagosomes plus autolysosomes was estimated from the number of mCherry-positive puncta, whereas puncta positive for GFP and mCherry were scored as autophagosomes APG.
Autophagic flux was calculated as the number of mCherry only fluorescent puncta autolysosomes, AL per cell. Before quantification, the maximum intensity projection was obtained for each cell image, and the cell and nucleus boundaries were extracted by respectively manual and automatic segmentation.
The fluorescence intensity density per ring was then measured and normalized with respect to the total intensity density of the cell see Appendix Fig.
S4 for details. At least 30 cells were measured for each condition. The xCELLigence system RTCA SP instrument Roche Applied Science monitors changes in the cell index a measure of cell attachment to the plate which has been shown to effectively correlate to proliferation, adhesion, and viability changes In no case was cell death due to excessive confluence as confirmed by plate inspection with a microscope.
To quantitatively measure apoptosis, the PE Annexin V Apoptosis Detection kit I BD-Bioscience was utilized. Samples were analyzed by flow cytometry on a BD FacsCalibur flow cytometer BD-Bioscience. To determine the apoptotic stage of the different cell populations, 7-AAD- and Annexin V-positive cells were determined with the CellQuest Pro Version 4.
Cells treated with staurosporine 2. Samples were analyzed by flow cytometry on a BD FacsCalibur flow cytometer BD-Bioscience , and data were analyzed with the FlowJo software. After several washes, cells were scraped and collected in the same buffer, centrifuged and the cell pellets were processed for embedding in the Epoxy, TAAB resin TAAB Laboratories according to standard procedures Autophagic vacuoles were identified using established criteria Autophagic vacuoles vesicles of 0.
Total autophagy vacuoles were composed of the sum of autophagosomes and autolysosomes. The maturation of autophagy vacuoles was calculated as the percent of autolysosomes and autophagosomes out of the total number of autophagy vacuoles.
Autophagic vacuoles were isolated from rat liver using discontinuous metrizamide density gradient Briefly, after homogenization, livers were subjected to sequential differential centrifugations to separate a fraction enriched in autophagy-related compartments and mitochondrial.
This fraction was placed at the bottom of a discontinuous three layers gradient of metrizamide and upon centrifugation, mitochondria remained at the bottom whereas a fraction enriched in autophagosomes is recovered in the top band and in autolysosomes in the second band from the top.
A lysosomal enriched fraction is also recovered from the third band of the gradient from the top. Upon extensive washing with 0. com or by the software Application Software Version 3. Flow cytometry data were analyzed with the FlowJo Software V7. Image J software v1. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
List of Figures that have associated raw data are Figs. Other data that support the findings of this study are available from the corresponding authors upon reasonable request. No datasets were generated or analyzed during this study. All unique materials generated are readily available from the authors.
Source data are provided with this paper. Kroemer, G. Autophagy and the integrated stress response. Cell 40 , — Article CAS PubMed PubMed Central Google Scholar.
Galluzzi, L. Linking cellular stress responses to systemic homeostasis. Cell Biol. Article CAS PubMed Google Scholar. Rybstein, M. The autophagic network and cancer.
Tasset, I. Role of chaperone-mediated autophagy in metabolism. FEBS J. Sciarretta, S. The role of autophagy in the heart.
et al. Molecular definitions of autophagy and related processes. EMBO J. Levine, B. Biological functions of autophagy genes: a disease perspective. Cell , 11—42 Klionsky, D. Guidelines for the use and interpretation of assays for monitoring autophagy 3rd edition.
Autophagy 12 , 1— Article PubMed PubMed Central Google Scholar. Yu, L. Autophagy termination and lysosome reformation regulated by mTOR. Nature , — Article ADS CAS PubMed PubMed Central Google Scholar. Füllgrabe, J.
The return of the nucleus: transcriptional and epigenetic control of autophagy. Article PubMed CAS Google Scholar. Laplante, M.
MTOR signaling in growth control and disease. Cell , — Saxton, R. mTOR signaling in growth, metabolism, and disease. Kim, J. mTOR as a central hub of nutrient signalling and cell growth. Sancak, Y. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids.
Article CAS Google Scholar. Condon, K. Nutrient regulation of mTORC1 at a glance. Cell Sci. Offermanns, S. Free fatty acid FFA and hydroxy carboxylic acid HCA receptors. Wauson, E. Minireview: nutrient sensing by G protein-coupled receptors.
Liu, S. Activation of Gαq in cardiomyocytes increases Vps34 activity and stimulates autophagy. Sánchez-Fernández, G. Gaq signalling: the new and the old.
Stolz, A. Cargo recognition and trafficking in selective autophagy. Cuervo, A. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Pei, Y. Engineered GPCRs as tools to modulate signal transduction. Article PubMed Google Scholar. Logie, L. Characterization of a protein kinase B inhibitor in vitro and in insulin-treated liver cells.
Bain, J. The selectivity of protein kinase inhibitors: a further update. Biochemical J. Article Google Scholar. Korolchuk, V. Lysosomal positioning coordinates cellular nutrient responses. Linares, J. Amino acid activation of mTORC1 by a PB1-domain-driven kinase complex cascade.
Cell Rep. Fan, G. Phospholipase C-independent activation of glycogen synthase kinase-3beta and C-terminal Src kinase by Galphaq. Shankaranarayanan, A.
Gαqallosterically activates and relieves autoinhibition of p63RhoGEF. Protein kinase C ζ interacts with a novel binding region of Gαq to act as functional effector protein.
Lamark, T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. Efeyan, A. Nutrients and growth factors in mTORC1 activation. Shimobayashi, M. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Sánchez-Martín, P.
Moscat, J. Cell 96, — CrossRef Full Text Google Scholar. Buttgereit, F. A hierarchy of ATP-consuming processes in mammalian cells. Castets, P. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy.
Chen, C. FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and Rictor. Cell 18, — Clemente-Postigo, M.
The role of autophagy in white adipose tissue function: implications for metabolic health. Metabolites Conza, D. The SGK1 inhibitor SI induces autophagy, apoptosis, and endoplasmic reticulum stress in endometrial cancer cells.
de la Calle Arregui, C. Limited survival and impaired hepatic fasting metabolism in mice with constitutive Rag GTPase signaling.
Efeyan, A. Nutrient-sensing mechanisms and pathways. Nature , — RagA, but not RagB, is essential for embryonic development and adult mice. Cell 29, — Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival.
Ezaki, J. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy 7, — Fejes-Tóth, G. Frescas, D. Nuclear trapping of the forkhead transcription factor FoxO1 via sirt-dependent deacetylation promotes expression of glucogenetic genes.
Fry, C. Skeletal muscle protein balance and metabolism in the elderly. Aging Sci. Ganley, I. Garami, A. Cell 11, — Hosokawa, N. Nutrient-dependent mTORC1 association with the ULK1-AtgFIP complex required for autophagy. Cell 20, — Hoxhaj, G.
The PI3K—AKT network at the interface of oncogenic signalling and cancer metabolism. Cancer 20, 74— Hsu, P. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling.
Inoki, K. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. Jewell, J. Differential regulation of mTORC1 by leucine and glutamine. Jia, R.
Lysosome positioning influences mTORC2 and AKT signaling. Cell 75, 26— Jiang, S. Karsli-Uzunbas, G. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. Kenerson, H. Tuberous sclerosis complex-1 deficiency attenuates diet-induced hepatic lipid accumulation.
PLoS One 6:e Kim, E. Regulation of TORC1 by Rag GTPases in nutrient response. Kim, J. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Komatsu, M. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice.
Korolchuk, V. Lysosomal positioning coordinates cellular nutrient responses. Kuma, A. The role of autophagy during the early neonatal starvation period.
Laplante, M. DEPTOR Cell-autonomously promotes adipogenesis, and its expression is associated with obesity. Lawrence, R. The lysosome as a cellular centre for signalling, metabolism and quality control. Lee, P. Liko, D. Loss of TSC complex enhances gluconeogenesis via upregulation of Dlk1-Dio3 locus miRNAs.
Liu, W. SGK1 inhibition-induced autophagy impairs prostate cancer metastasis by reversing EMT. Cancer Res. Magdalon, J. Constitutive adipocyte mTORC1 activation enhances mitochondrial activity and reduces visceral adiposity in mice.
Acta Mol. Lipids , — Mammucari, C. FoxO3 controls autophagy in skeletal muscle in vivo. Martina, J. Rag GTPases mediate amino acid-dependent recruitment of TFEB and MITF to lysosomes.
Masiero, E. Autophagy is required to maintain muscle mass. Medina, D. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB.
Mutvei, A. Rap1-GTPases control mTORC1 activity by coordinating lysosome organization with amino acid availability. Napolitano, G. TFEB at a glance. Cell Sci. A substrate-specific mTORC1 pathway underlies Birt—Hogg—Dubé syndrome. MXL-3 and HLH transcriptionally link lipolysis and autophagy to nutrient availability.
Ortega-Molina, A. Oncogenic Rag GTPase signalling enhances B cell activation and drives follicular lymphoma sensitive to pharmacological inhibition of mTOR. Park, Y. mTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4.
Peng, M. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to Control mTORC1 signaling. Cell , — Peterson, T. mTOR complex 1 regulates lipin 1 localization to control the srebp pathway.
Pyo, J. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Rabanal-Ruiz, Y. mTORC1 and nutrient homeostasis: The central role of the lysosome. Robitaille, A. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis.
Roczniak-Ferguson, A. The Transcription Factor TFEB Links mTORC1 signaling to transcriptional control of lysosome homeostasis. Rogala, K. Structural basis for the docking of mTORC1 on the lysosomal surface. Sancak, Y. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1.
Sandri, M. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Sardiello, M. A gene network regulating lysosomal biogenesis and function. Saxton, R. mTOR signaling in growth, metabolism, and disease.
Schalm, S. Identification of a conserved motif required for mTOR signaling. Schiaffino, S. Autophagic degradation of glycogen in skeletal muscles of the Newborn rat. Schurmann, A. Cloning of a novel family of mammalian GTP-binding proteins RagA, RagBs, RagB1 with remote similarity to the Ras-related GTPases.
Sengupta, S. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Settembre, C. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. TFEB links autophagy to lysosomal biogenesis.
Shimobayashi, M. Multiple amino acid sensing inputs to mTORC1. Cell Res. Singh, R. Autophagy regulates lipid metabolism. Autophagy regulates adipose mass and differentiation in mice. Stitt, T.
Cell 14, — Tee, A. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Torrence, M. The mTORC1-mediated activation of ATF4 promotes protein and glutathione 1 synthesis downstream of growth signals 2.
bioRxiv [Preprint]. Valvezan, A. Molecular logic of mTORC1 signalling as a metabolic rheostat. mTORC1 couples nucleotide synthesis to nucleotide demand resulting in a targetable metabolic vulnerability.
Cancer Cell 32, — Wada, S. The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Wulff, P. Yang, L. Defective hepatic autophagy in obesity promotes er stress and causes insulin resistance.
Yecies, J. Akt stimulates hepatic SREBP1c and lipogenesis through Parallel mTORC1-dependent and independent pathways. Yu, L. Termination of autophagy and reformation of lysosomes regulated by mTOR.
Yu, Y. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Zhang, Y. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Adipose-specific deletion of autophagy-related gene 7 atg7 in mice reveals a role in adipogenesis.
Coordinated regulation of protein synthesis and degradation by mTORC1. Zhao, J. Zhao, Y. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity.
Zhou, B.
The mechanistic sugnaling of Autophagy and mTOR signaling mTORmaster regulator of cellular metabolism, exists in two distinct Autophagy and mTOR signaling mTOR complex Auophagy and mTOR Autophgy 2 mTORC1 and signaliny. MTORC1 is a master switch for most energetically Fuel Management Software processes in Autophagy and mTOR signaling cell, driving cell growth and building cellular biomass in instances of nutrient sivnaling, and conversely, allowing autophagic recycling of cellular components upon nutrient limitation. The means by which the mTOR kinase blocks autophagy include direct inhibition of the early steps of the process, and the control of the lysosomal degradative capacity of the cell by inhibiting the transactivation of genes encoding structural, regulatory, and catalytic factors. Upon inhibition of mTOR, autophagic recycling of cellular components results in the reactivation of mTORC1; thus, autophagy lies both downstream and upstream of mTOR. The functional relationship between the mTOR pathway and autophagy involves complex regulatory loops that are significantly deciphered at the cellular level, but incompletely understood at the physiological level.Autophagy and mTOR signaling -
View PDF. To show local product price and availability and for ordering, we are taking you now to our secure CST Portal.
Would you like to visit your country specific website? YES NO. Save This Selection. All Rights Reserved. Pathway Description Legend Pathway Description: Macroautophagy, often referred to as autophagy, is a catabolic process that results in the autophagosomic-lysosomal degradation of bulk cytoplasmic contents, abnormal protein aggregates, and excess or damaged organelles.
Codogno P, Mehrpour M, Proikas-Cezanne T Canonical and non-canonical autophagy: variations on a common theme of self-eating? Cell Biol. Ding WX, Yin XM Mitophagy: mechanisms, pathophysiological roles, and analysis.
Feng D, Liu L, Zhu Y, Chen Q Molecular signaling toward mitophagy and its physiological significance. Cell Res. Papinski D, Kraft C Atg1 kinase organizes autophagosome formation by phosphorylating Atg9.
Autophagy 10 7 , — Schneider JL, Cuervo AM Autophagy and human disease: emerging themes. created September revised September Request Permission for Pathway View PDF.
Learn More With PhosphoSite Plus ®. Image on the right shows the quantification analysis of lysosome distribution. Representative blots of three independent experiments are shown.
Control vs. Immunofluorescence with LAMP1 to highlight the endolysosomal compartment revealed that WT MEFs displayed the expected switch of lysosomes from the cell periphery to a more perinuclear location upon exposure to amino acid-free medium 25 , whereas the re-exposure to amino acids restored their original location Fig.
These results are consistent with their inability to stimulate mTORC1 in response to nutrient recovery. Intriguingly, immunoprecipitation of either endogenous Gαq Fig. Importantly, immunoprecipitation of endogenous Raptor, a specific component of mTORC1, also revealed the presence in the same complex of Gαq, p62, and mTOR Fig.
The mTORC1 activation complex formation undergoes a dynamic modulation upon nutrient fluctuations 15 , Following amino acid deprivation, we found dissociation of p62 from mTOR, while Raptor and Gαq remained in the mTOR complex Fig.
In response to amino acid replenishment, p62 re-associated to the mTORC1 complex. Blots are representative of three a , c and four b independent experiments. c WT vs. d Control vs. We searched for potential molecular links between Gαq and mTORC1 activation. A novel acidic region in Gαq responsible for its direct interaction with the PB1-containing protein Protein kinase C ζ PKCζ has been reported, and glutamic acid residues and proven to be critical for such interaction Remarkably, the stimulation of the mTORC1 pathway triggered by transient expression of Gαq WT in low-serum conditions was not mimicked upon overexpression of the Gq-EEAA mutant Fig.
Overall, these results suggested that the PB1-binding region of Gαq is involved in mTORC1 pathway modulation. A representative blot of three independent experiments is shown. c — f CHO cells were transiently transfected with the indicated combinations of HA-p62, Gαq WT or the indicated Gαq mutants constitutively active Gq-QL or GqRC, active but PLC-β activation-defective GqQL-AA, defective in binding to PB1-domain-containing proteins Gq-EEAA and constitutively active GqRC-EEAA , or the PB1 domain of p62 GFP-Flag-PB1-p Gαq WT vs.
We, therefore, used transient overexpression of Gαq and HA-p62 constructs to characterize their association. Of note, p62 displays features of a bona fide Gαq effector reviewed in ref.
Consistent with data in Fig. Of note, Gαq mutants Gαq-YF, Gαq-TE, Gαq-WD unable to bind GRK2 co-immunoprecipitated with p62 to an even greater extent than wild-type Gαq Supplementary Fig.
Consistent with a PB1-like association between Gαq and p62, the overexpression of the PB1 domain of p62 clearly inhibited the complex formation Fig. a CHO cells were transfected with HA-p62 and Gαq constructs, starved with 0.
c Basal activation status of the mTORC1 cascade and autophagy levels correlates with the ability of Gαq mutants to interact with p mTOR immunoprecipitates and total lysates were analyzed by western blot with specific antibodies to determine the components of the complex and overall cell expression levels, respectively.
The blots shown are representative of three independent experiments. e mTORC1 reactivation and autophagy modulation in response to amino acid recovery after starvation correlate with the ability of Gαq to interact with p Statistical significance was analyzed using two-way ANOVA with Bonferroni multiple comparison test.
Basal activation status of the mTORC1 cascade and autophagy levels correlated with the ability of Gαq mutants to interact with p Overall, these results support the notion that the interaction between Gαq and p62 in response to nutrients is key for the stimulation of the mTORC1 pathway and the control of autophagy.
Our data suggest that Gαq acts as a general and relevant modulator of mTORC1 signaling over autophagy in response to fluctuations in different types of nutrients.
The metabolic modulation of autophagy is mainly orchestrated by the balance between the opposite inputs provided by the mTORC1 negative and AMPK positive pathways 12 , 31 , Moreover, constitutively active mutations in Gαq maintain mTORC1 activated in absence of serum.
Fifth, consistent with its role as an autophagy modulator, we demonstrate the presence of an endogenous Gαq pool in autophagic compartments and lysosomes in rat liver, where it co-localizes with mTOR, Raptor, and p62, and immunoprecipitation of endogenous mTOR or Raptor from WT MEFs in the presence of nutrients shows that Gαq is part of the mTORC1 multimolecular complex.
This signaling adaptor contains several functional regions and displays a variety of subcellular locations, including autophagosomes and lysosomes reviewed in refs. p62 plays a central role as an autophagy receptor by forming PB1-linked homo-oligomers, recruiting ubiquitinated cargos, and inducing nucleation of the autophagosome membrane via its interaction with LC3 33 being itself a substrate of autophagy p62 is also involved in other cellular processes 33 , including the modulation of mTORC1 see below.
The mTORC1 signaling hub displays an intricate spatiotemporal regulation, involving both upstream signaling pathways and timely recruitment to lysosomes 12 , Major components of the mTORC1 complex include mTOR kinase, mLST8, DEPTOR, and the specific regulatory protein Raptor, which plays a relevant role in facilitating substrate recruitment and in controlling mTORC1 subcellular localization 12 , Importantly, the presence of amino acids, acting through different sensors, is key for full mTORC1 activation by facilitating its recruitment to the lysosomal membrane.
This is achieved via the Ras-related GTP-binding protein Rag family of small GTPases. Glucose-dependent mTORC1 stimulation is also partially regulated through Rag-GTPases by complex mechanisms reviewed in refs.
Thus, Rags appear to integrate multiple nutrient inputs converging in mTORC1 modulation. Importantly, in the presence of amino acids, p62 has been reported to interact with Raptor, thus helping mTORC1 complex translocation to the lysosome The presence of p62 in the mTORC1 multimolecular complex might also contribute to the full activation of kinase activity via TRAF6-mediated K63 ubiquitination of mTOR First, p62, mTOR, Raptor, and Gαq are present in the same multimolecular complex in endogenous conditions in the presence of nutrients, whereas cells lacking Gαq display a reduced presence of p62 in mTOR and Raptor immunoprecipitates along with decreased mTORC1 pathway activation.
Gαq and p62 appear to associate through a PB1-like interaction, involving the PB1 domain of p62 and the acidic PB1-binding region of Gαq, different from the specific interface involved in the association of Gαq with the canonical PLCβ or p63RhoGEF effectors This acidic Gαq region has been shown to interact with the PB1 domain of PKCζ 29 or with the cold-activated channel TRPM8 42 , consistent with this Gαq region being a functional module.
Such core Gαq scaffolding role would be consistent with its general role in autophagy control in response to different types of nutrient stress see scheme in Fig. In nutrient-sufficiency conditions, the presence of serum, amino acids, or glucose would foster the PB1-like interaction between Gαq and p62, leading to basal mTORC1 stimulation and preservation of low, homeostatic autophagy levels.
Upper part: In nutrient-sufficiency conditions, the presence of serum, amino acids, or glucose would foster the PB1-like interaction between Gαq and p62, leading to basal mTORC1 stimulation and preservation of homeostatic autophagy levels.
Lower part: Autophagy levels diagram. In response to starvation, there is a controlled and transient upregulation of autophagy, with a subsequent return to basal levels upon the partial recovery of nutrients.
An increasing number of nutrients and metabolites are being identified as endogenous ligands for Gq-coupled GPCR 16 , 17 , On the other hand, relevant serum components as lysophosphatidic acid or sphingosinephosphate and factors as insulin or IGF-1 have been reported to directly stimulate Gαq signaling The presence of Gαq in lysosomal and autophagy-related organelles may be subsequent to receptor-mediated activation at the plasma membrane or represent a stable pool able to undergo nutrient-mediated stimulation at such locations.
Recent data indicate the presence of Gαq in endosomes and other organelles, such as mitochondria 49 , The agonist-dependent presence of GPCR and heterotrimeric G proteins in endosomes as a consequence of GPCR recycling or degradation mechanisms has also been reported Since endocytosis-derived organelles, can form elongation membranes to finally form autophagosomes in response to starvation 54 , endosomes might constitute a source for Gαq in autophagy-related organelles.
Alternatively, activation of Gαq by nutrient-sensitive GPCR could take place directly at locations different from the plasma membrane via molecules that can gain access to the intracellular milieu via specific transporters, as is the case for amino acids, glucose, and other nutrients.
In sum, our results postulate Gαq as a key hub in the autophagy machinery. All rats were housed under pathogen-free conditions and handled according to protocols approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine New York, USA.
Since for these experiments mice were not treated with tamoxifen, we refer to them as WT mice as they display endogenous Gαq levels. We did not use randomization in our animal studies. We were not blinded to the group in our animal studies.
Offermanns Max-Planck-Institute for Heart and Lung Research, Germany. Atg5 WT and Atg5 KO MEFs were provided by Dr. Mizushima University of Tokyo, Tokyo, Japan and DREADD-Gq-HEK cells human female by Dr.
Silvio Gutkind University of San Diego, California, USA CHO cells overexpressing the muscarinic M3 acetylcholine receptor CHO-M3 mice female were a kind gift from Dr.
Tobin University of Glasgow, UK. Cell lines were authenticated by short tandem repeat STR or DNA barcoding analysis and tested for mycoplasma contamination.
Human umbilical vein endothelial cells HUVEC human male were grown in Medium Life Technologies, without NaHCO 3 supplemented with 2.
Explants were homogenized using metal beads in a Tissue Lyser Qiagen, Hilden, Germany 60 , and lysates used for immunoblot analysis of mTORC1 pathway modulation. For autophagy modulation by nutrient depletion assays, cells were maintained in 0. Nutrient recovery experiments in MEF cells were carried out by adding increasing percentages of bovine serum or a full amino acid mix MEM amino acids solution 50× Sigma-Aldrich.
For degradation assays, cells were treated with the lysosomal inhibitors mentioned and the proteasomal inhibitor MG 0. The Gαq and Gαq-RC cDNA were from our laboratories Dr. Aragay, CSIC, Barcelona, Spain. Lin Stony Brook University, New York, USA. The Gαq mutant proteins that lack the ability to interact with GRK2 Gαq-TE, WD, YF were a gift from Dr.
Kozasa The University of Tokyo, Japan. GRK2, GRK2-KR, GRK2-DA cDNAs were a gift from Dr. Benovic The Kimmel Cancer Center, USA. The GRK2-RH domain cDNA 61 was generated in our laboratory. The HA-p62 construct was provided by Dr.
Campbell University of Illinois at Chicago, USA. GFP-Flag-PB1-p62 was provided by Dr. Berk University of Rochester, NY, USA. mCherry-GFP-LC3 Addgene, Dr. Mizushima, constructed by T. Kaizuka University of Tokyo, Japan. cDNAs for lentiviral construction pMD2. G, pSD. Iggo University of St Andrews, Edinburgh, UK.
Gq-wt and psd Gq-EEAA were created in our laboratory from psd GqQL Addgene, Dr. All primers' information has been included in Supplementary Table 1. Gq-EEAA DNA constructs were transfected together with psPAX2 and pMD2. All generated MEF cells lines, psd. Cells were seeded in a six-well plate a day before transduction.
Then, a 2× polybrene mixture was prepared by adding 0. Depending on the cell type these stable cell lines were used until passage 3—4. Immunoprecipitation was performed with agarose-conjugated anti-HA antibodies Santa Cruz, F-7, scAC.
Endogenous immunoprecipitation assays were performed by incubating 0. Protein concentration was measured using the DC Protein Assay Reagents A, B, and S Bio-Rad. Immunoreactivity bands were quantified by laser densitometry with a Biorad GS scanner and using de Bio-Rad provided Image Lab 5. Vertical lines in western blots indicate juxtaposed lanes that come from the same gel but were nonadjacent.
Cells were mounted with Fluoromount-G-Dapi Southern Biotech. Sample images were acquired using a confocal laser microscope LSM Zeiss at ×63 magnification. Image J software NIH was used for general image analysis and manipulations. Imaging of the mCherry-GPF-LC3 reporter was carried out using high-content microscopy.
The total number of autophagic vacuoles AV, autophagosomes plus autolysosomes was estimated from the number of mCherry-positive puncta, whereas puncta positive for GFP and mCherry were scored as autophagosomes APG.
Autophagic flux was calculated as the number of mCherry only fluorescent puncta autolysosomes, AL per cell. Before quantification, the maximum intensity projection was obtained for each cell image, and the cell and nucleus boundaries were extracted by respectively manual and automatic segmentation.
The fluorescence intensity density per ring was then measured and normalized with respect to the total intensity density of the cell see Appendix Fig. S4 for details. At least 30 cells were measured for each condition. The xCELLigence system RTCA SP instrument Roche Applied Science monitors changes in the cell index a measure of cell attachment to the plate which has been shown to effectively correlate to proliferation, adhesion, and viability changes In no case was cell death due to excessive confluence as confirmed by plate inspection with a microscope.
To quantitatively measure apoptosis, the PE Annexin V Apoptosis Detection kit I BD-Bioscience was utilized. Samples were analyzed by flow cytometry on a BD FacsCalibur flow cytometer BD-Bioscience. To determine the apoptotic stage of the different cell populations, 7-AAD- and Annexin V-positive cells were determined with the CellQuest Pro Version 4.
Cells treated with staurosporine 2. Samples were analyzed by flow cytometry on a BD FacsCalibur flow cytometer BD-Bioscience , and data were analyzed with the FlowJo software. After several washes, cells were scraped and collected in the same buffer, centrifuged and the cell pellets were processed for embedding in the Epoxy, TAAB resin TAAB Laboratories according to standard procedures Autophagic vacuoles were identified using established criteria Autophagic vacuoles vesicles of 0.
Total autophagy vacuoles were composed of the sum of autophagosomes and autolysosomes. The maturation of autophagy vacuoles was calculated as the percent of autolysosomes and autophagosomes out of the total number of autophagy vacuoles.
Autophagic vacuoles were isolated from rat liver using discontinuous metrizamide density gradient Briefly, after homogenization, livers were subjected to sequential differential centrifugations to separate a fraction enriched in autophagy-related compartments and mitochondrial.
This fraction was placed at the bottom of a discontinuous three layers gradient of metrizamide and upon centrifugation, mitochondria remained at the bottom whereas a fraction enriched in autophagosomes is recovered in the top band and in autolysosomes in the second band from the top.
A lysosomal enriched fraction is also recovered from the third band of the gradient from the top. Upon extensive washing with 0. com or by the software Application Software Version 3. Flow cytometry data were analyzed with the FlowJo Software V7.
Image J software v1. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
List of Figures that have associated raw data are Figs. Other data that support the findings of this study are available from the corresponding authors upon reasonable request. No datasets were generated or analyzed during this study.
All unique materials generated are readily available from the authors. Source data are provided with this paper. Kroemer, G. Autophagy and the integrated stress response. Cell 40 , — Article CAS PubMed PubMed Central Google Scholar. Galluzzi, L. Linking cellular stress responses to systemic homeostasis.
Cell Biol. Article CAS PubMed Google Scholar. Rybstein, M. The autophagic network and cancer. Tasset, I. Role of chaperone-mediated autophagy in metabolism. FEBS J. Sciarretta, S.
The role of autophagy in the heart. et al. Molecular definitions of autophagy and related processes. EMBO J. Levine, B. Biological functions of autophagy genes: a disease perspective. Cell , 11—42 Klionsky, D. Guidelines for the use and interpretation of assays for monitoring autophagy 3rd edition.
Autophagy 12 , 1— Article PubMed PubMed Central Google Scholar. Yu, L. Autophagy termination and lysosome reformation regulated by mTOR. Nature , — Article ADS CAS PubMed PubMed Central Google Scholar.
Füllgrabe, J. The return of the nucleus: transcriptional and epigenetic control of autophagy. Article PubMed CAS Google Scholar. Laplante, M. MTOR signaling in growth control and disease. Cell , — Saxton, R. mTOR signaling in growth, metabolism, and disease. Kim, J.
mTOR as a central hub of nutrient signalling and cell growth. Sancak, Y. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Article CAS Google Scholar. Condon, K. Nutrient regulation of mTORC1 at a glance.
Cell Sci. Offermanns, S. Free fatty acid FFA and hydroxy carboxylic acid HCA receptors. Wauson, E. Minireview: nutrient sensing by G protein-coupled receptors.
Liu, S. Activation of Gαq in cardiomyocytes increases Vps34 activity and stimulates autophagy. Sánchez-Fernández, G. Gaq signalling: the new and the old.
Stolz, A. Cargo recognition and trafficking in selective autophagy. Cuervo, A. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation.
Pei, Y. Engineered GPCRs as tools to modulate signal transduction. Article PubMed Google Scholar. Logie, L. Characterization of a protein kinase B inhibitor in vitro and in insulin-treated liver cells.
Bain, J. The selectivity of protein kinase inhibitors: a further update. Biochemical J. Article Google Scholar. Korolchuk, V. Lysosomal positioning coordinates cellular nutrient responses. Linares, J. Amino acid activation of mTORC1 by a PB1-domain-driven kinase complex cascade.
Cell Rep. Fan, G. Phospholipase C-independent activation of glycogen synthase kinase-3beta and C-terminal Src kinase by Galphaq. Shankaranarayanan, A. Gαqallosterically activates and relieves autoinhibition of p63RhoGEF. Protein kinase C ζ interacts with a novel binding region of Gαq to act as functional effector protein.
Lamark, T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. Efeyan, A. Nutrients and growth factors in mTORC1 activation. Shimobayashi, M. Making new contacts: the mTOR network in metabolism and signalling crosstalk.
Sánchez-Martín, P. Moscat, J. p62 in cancer: signaling adaptor beyond autophagy. Ichimura, Y. Selective degradation of p62 by autophagy.
Lawrence, R. The lysosome as a cellular centre for signalling, metabolism and quality control. A nutrient-induced affinity switch controls mTORC1 activation by its Rag GTPase—ragulator lysosomal scaffold. Yang, G. RagC phosphorylation autoregulates mTOR complex 1.
Tan, V. Nutrient-sensing mTORC1: integration of metabolic and autophagic signals. Duran, A. P62 is a key regulator of nutrient sensing in the mTORC1 pathway. Cell 44 , — K63 polyubiquitination and activation of mTOR by the pTRAF6 complex in nutrient-activated cells.
Cell 51 , — Zhang, X. Direct inhibition of the cold-activated TRPM8 ion channel by G α q. Amino acid regulation of autophagy through the GPCR TAS1R1-TAS1R3.
Autophagy 9 , — Mercan, F. Novel role for SHP-2 in nutrient-responsive control of S6 kinase 1 signaling. Khan, M. Role of inositol trisphosphate receptors in autophagy in DT40 cells. Manning, B. Husted, A.
GPCR-mediated signaling of metabolites. Cell Metab.
Thank you for visiting nature. You are using a browser version with signalin support for CSS. To nad the Autophagy and mTOR signaling experience, we recommend Sports dietary analysis use Autophagy and mTOR signaling more up to date browser or turn aand compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. The mTORC1 node plays a major role in autophagy modulation. We report a role of the ubiquitous Gαq subunit, a known transducer of plasma membrane G protein-coupled receptors signaling, as a core modulator of mTORC1 and autophagy. They are also unable to reactivate mTORC1 and thus inactivate ongoing autophagy upon nutrient recovery. Autophagy and mTOR signaling SLProud CGWyttenbach Signalibg. mTOR's role Autophaagy ageing: protein synthesis Autophayg autophagy?. Autophagy and mTOR signaling Albany NY. Copyright: signalign Hands et al. This is an Autophayy article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. The molecular and cellular mechanisms that regulate ageing are currently under scrutiny because ageing is linked to many human diseases. The nutrient sensing TOR pathway is emerging as a key regulator of ageing.
Nach meiner Meinung lassen Sie den Fehler zu. Geben Sie wir werden es besprechen. Schreiben Sie mir in PM, wir werden reden.
Nach meiner Meinung irren Sie sich. Ich biete es an, zu besprechen. Schreiben Sie mir in PM, wir werden umgehen.
Eben dass wir ohne Ihre ausgezeichnete Phrase machen würden
Sie sind nicht recht. Geben Sie wir werden es besprechen. Schreiben Sie mir in PM, wir werden umgehen.